Title
题目
Robust image representations with counterfactual contrastive learning
基于反事实对比学习的鲁棒图像表征
01
文献速递介绍
医学影像中的对比学习已成为利用未标记数据的有效策略。这种自监督学习方法已被证明能显著提升模型跨领域偏移的泛化能力,并减少训练所需的高质量标注数据量(Azizi等人,2021,2023;Ghesu等人,2022;Zhou等人,2023)。然而,基于对比的学习能否成功在很大程度上依赖于正样本对生成流程(Tian等人,2020)。这些正样本对通常通过对原始图像重复应用预定义的数据增强生成。因此,增强流程的变化会对所学表征的质量产生显著影响,最终影响下游任务性能以及对领域变化的鲁棒性(Tian等人,2020;Scalbert等人,2023)。传统上,为自然图像开发的增强流程被直接应用于医学影像,但由于医学影像采集方式存在独特挑战和特点,这种做法可能并非最优。特别是,领域变异通常远大于细微的类别差异。这可能导致通过对比学习获得的表征无意中将这些无关的采集相关变异编码到所学表征中。 在本研究中,我们旨在提高通过对比学习获得的表征对领域偏移(尤其是采集偏移)的鲁棒性。采集偏移由图像采集协议的变化(设备设置、后处理软件等)引起,是医学影像领域数据集偏移的主要来源。我们假设,通过在正样本对创建阶段更真实地模拟领域变异,可以提高对比学习特征对这类图像特征变化的鲁棒性。为此,我们提出并评估了“反事实对比学习”——一种新的对比样本对生成框架,它利用深度生成模型的最新进展来生成高质量、真实的反事实图像(Ribeiro等人,2023;Fontanella等人,2023)。反事实生成模型使我们能够回答“如果……会怎样”的问题,例如模拟用一种设备采集的乳腺X线影像若用另一种设备采集会呈现何种样子。具体而言,在我们提出的反事实对比框架中,我们通过将真实图像与其领域反事实图像匹配,创建跨领域正样本对,从而真实模拟设备变化。重要的是,所提出的方法不依赖于对比目标的选择,因为它仅影响正样本对的创建步骤。我们以两种广泛使用的对比学习框架为例阐述该方法的优势:开创性研究SimCLR(Chen等人,2020)和新发布的DINO-V2(Oquab等人,2023)目标。此外,为精确衡量所提出的反事实样本对生成过程的效果,我们还将该方法与一种更简单的方法进行对比——后者仅通过生成的反事实图像扩展训练集。 我们在两种医学影像模态(乳腺X线和胸部X线)、五个公共数据集以及两项临床相关分类任务上评估所提出的反事实对比学习框架,结果表明我们的方法能生成对领域变化更鲁棒的特征。这种增强的鲁棒性在特征空间中可直接观察到,更重要的是,它带来了下游任务性能的显著提升,尤其是在标签有限的情况下以及针对训练时代表性不足的领域。关键的是,尽管训练目标存在显著差异,这些发现对SimCLR和DINO-V2均成立。本文是我们近期MICCAI研讨会论文(Roschewitz等人,2024)的扩展。其差异体现在以下方面: • 我们此前仅考虑了SimCLR目标,本文将反事实对比方法扩展至新提出的DINO-V2(Oquab等人,2023)目标。这些新结果从经验上证明,所提出的方法具有通用性,且不依赖于对比目标的选择。 • 尽管本研究的主要焦点是对采集偏移的鲁棒性,但在本扩展中,我们表明所提出的方法可扩展至其他场景,例如改善亚组性能。 • 我们大幅扩充了讨论、方法和相关工作部分。
Abatract
摘要
Contrastive pretraining can substantially increase model generalisation and downstream performance. However, the quality of the learned representations is highly dependent on the data augmentation strategy appliedto generate positive pairs. Positive contrastive pairs should preserve semantic meaning while discardingunwanted variations related to the data acquisition domain. Traditional contrastive pipelines attempt tosimulate domain shifts through pre-defined generic image transformations. However, these do not alwaysmimic realistic and relevant domain variations for medical imaging, such as scanner differences. To tackle thisissue, we herein introduce counterfactual contrastive learning, a novel framework leveraging recent advances incausal image synthesis to create contrastive positive pairs that faithfully capture relevant domain variations.Our method, evaluated across five datasets encompassing both chest radiography and mammography data,for two established contrastive objectives (SimCLR and DINO-v2), outperforms standard contrastive learningin terms of robustness to acquisition shift. Notably, counterfactual contrastive learning achieves superiordownstream performance on both in-distribution and external datasets, especially for images acquired withscanners under-represented in the training set. Further experiments show that the proposed framework extendsbeyond acquisition shifts, with models trained with counterfactual contrastive learning reducing subgroupdisparities across biological sex.
对比预训练能显著提升模型的泛化能力和下游任务性能。然而,所学表征的质量高度依赖于用于生成正样本对的数据增强策略。对比学习中的正样本对应在保留语义含义的同时,摒弃与数据采集领域相关的非期望变异。传统的对比学习流程试图通过预定义的通用图像变换来模拟领域偏移。但对于医学影像而言,这些变换并不总能模拟真实且相关的领域变异,例如扫描仪差异。 为解决这一问题,我们在此提出反事实对比学习——一种利用因果图像合成最新进展的新型框架,旨在生成能真实捕捉相关领域变异的对比正样本对。我们的方法在五个涵盖胸部X线和乳腺X线数据的数据集上进行了评估,针对两种成熟的对比目标(SimCLR和DINO-v2),结果显示其在对采集偏移的鲁棒性方面优于标准对比学习。值得注意的是,反事实对比学习在分布内数据集和外部数据集上均实现了更优的下游性能,尤其对于训练集中代表性不足的扫描仪所采集的图像而言。进一步实验表明,该框架的优势不仅限于采集偏移:经反事实对比学习训练的模型还能减少不同生物学性别亚组间的差异。
Results
结果
In this section, we compare the quality and robustness of thelearned representations for various pre-training paradigms. First, standard SimCLR. Secondly, SimCLR+, where we train a model using classicSimCLR on a training set enriched with domain counterfactuals. Finally,CF-SimCLR combines SimCLR with our proposed counterfactual contrastive pair generation framework. We then repeat the same analysisfor models pre-trained with the DINO objective, comparing DINO, CFDINO and DINO+. Note that in SimCLR+ (resp. DINO+), counterfactualsand real images are not paired during the contrastive learning step;they are all considered as independent training samples. As such, SimCLR+/DINO+ represent the common paradigm of simply enriching thetraining set with synthetic examples. In CF-SimCLR/CF-DINO, on theother hand, we systematically pair real images with their correspondingcounterfactual for positive pair creation (Fig. 1).We compare the effect of these three pretraining strategies on chestX-rays and mammograms. For chest X-rays, we evaluate the qualityof the learned representations by assessing downstream performanceon pneumonia detection. For mammography, we focus on the task ofbreast density prediction (important for risk modelling). Pre-trainingstrategies are evaluated with linear probing (i.e. classifiers trained ontop of frozen encoders) as well as full model finetuning (unfrozen encoders). Linear probes are best representative of the quality of learnedrepresentation during pre-training, as representations are unchangedduring downstream training. However we also include the comparisonwith full model finetuning, as this is often the training paradigm ofchoice in practical scenarios. All models are finetuned with real dataonly, using a weighted cross-entropy loss. We evaluate the pre-trainedencoders in two settings. First, we test the encoders on ID datasets,i.e. using the same data for pre-training, finetuning and testing. Secondly, encoders are evaluated on OOD datasets, i.e. where the model isfinetuned/linear-probed and tested on data external to the pre-trainingdata. Evaluation on external datasets is crucial to assess how counterfactual contrastive pretraining performs on unseen domains (outside ofscanner distribution used for training the causal inference model). Allencoders are pretrained on the full, unlabelled, PadChest and EMBEDdatasets. However, the main motivation for self-supervised pretrainingis to increase robustness when only a limited amount of labelled datais available (Azizi et al., 2023). Hence, we evaluate the encoders forvarying amounts of annotated data. Specifically, we finetune (or linearprobe) the encoder using a pre-defined amount of labelled samples, andthen evaluate the resulting classifier on a fixed test set. We repeat thisprocess several times, varying the amount of labelled samples to assessthe effect of pre-training in function of number of labelled samplesavailable for training the downstream classifier, e.g. for PadChest, thenumber of labelled training samples varies from 3249 to 64,989. Allour code is publicly available at https://github.com/biomedia-mira/counterfactual-contrastive.For each task, we compare downstream performance across variouspretraining strategies, for each scanner, for both SimCLR (Figs. 5 and
and DINO (Figs. 9 and 11) objectives. Moreover, to help visualiseperformance differences across encoders, we report the performancedifferences between the proposed encoders and the baseline in Figs.6, 8, 10 and 12.
在本节中,我们对比了不同预训练范式下所学表征的质量和鲁棒性。首先是标准的SimCLR;其次是SimCLR+,即在添加了领域反事实图像的训练集上,使用经典SimCLR方法训练模型;最后是CF-SimCLR,它将SimCLR与我们提出的反事实对比样本对生成框架相结合。随后,我们对采用DINO目标函数预训练的模型进行了相同分析,对比了DINO、CF-DINO和DINO+。需要注意的是,在SimCLR+(分别对应DINO+)中,反事实图像与真实图像在对比学习步骤中并未配对,而是均被视为独立的训练样本。因此,SimCLR+/DINO+代表了一种常见范式——仅通过合成样本来扩充训练集。与之不同的是,在CF-SimCLR/CF-DINO中,我们系统性地将真实图像与其对应的反事实图像配对,用于正样本对的创建(图1)。 我们在胸部X线和乳腺X线影像上对比了这三种预训练策略的效果。对于胸部X线,我们通过评估肺炎检测下游任务的性能来衡量所学表征的质量;对于乳腺X线,我们重点关注乳腺密度预测任务(这对风险建模至关重要)。预训练策略的评估采用两种方式:线性探测(即在冻结的编码器顶部训练分类器)和全模型微调(解冻编码器)。线性探测最能代表预训练阶段所学表征的质量,因为在下游训练过程中表征保持不变。但我们也纳入了与全模型微调的对比,因为这通常是实际场景中首选的训练范式。所有模型均仅使用真实数据进行微调,并采用加权交叉熵损失函数。 我们在两种设置下评估预训练编码器:首先,在分布内(ID)数据集上测试编码器,即预训练、微调和测试均使用相同数据;其次,在分布外(OOD)数据集上评估编码器,即模型的微调/线性探测和测试均使用预训练数据之外的数据。在外部数据集上的评估至关重要,这能衡量反事实对比预训练在未见过的领域(超出用于训练因果推断模型的扫描仪分布范围)上的表现。所有编码器均在完整的、未标记的PadChest和EMBED数据集上进行预训练。 然而,自监督预训练的主要目的是在仅有有限标注数据可用时提高模型鲁棒性(Azizi等人,2023)。因此,我们在不同数量的标注数据下对编码器进行了评估。具体而言,我们使用预定义数量的标注样本对编码器进行微调(或线性探测),然后在固定测试集上评估所得分类器的性能。我们重复这一过程多次,通过改变标注样本的数量,来评估预训练效果随下游分类器训练可用标注样本数量的变化——例如,在PadChest数据集中,标注训练样本的数量从3249个到64989个不等。我们的所有代码均公开可获取,链接为:https://github.com/biomedia-mira/counterfactual-contrastive。 对于每个任务,我们在SimCLR(图5和图7)和DINO(图9和图11)两种目标函数下,按扫描仪分别对比了不同预训练策略的下游任务性能。此外,为便于可视化不同编码器之间的性能差异,我们在图6、图8、图10和图12中报告了所提编码器与基线模型的性能差异。
Figure
图
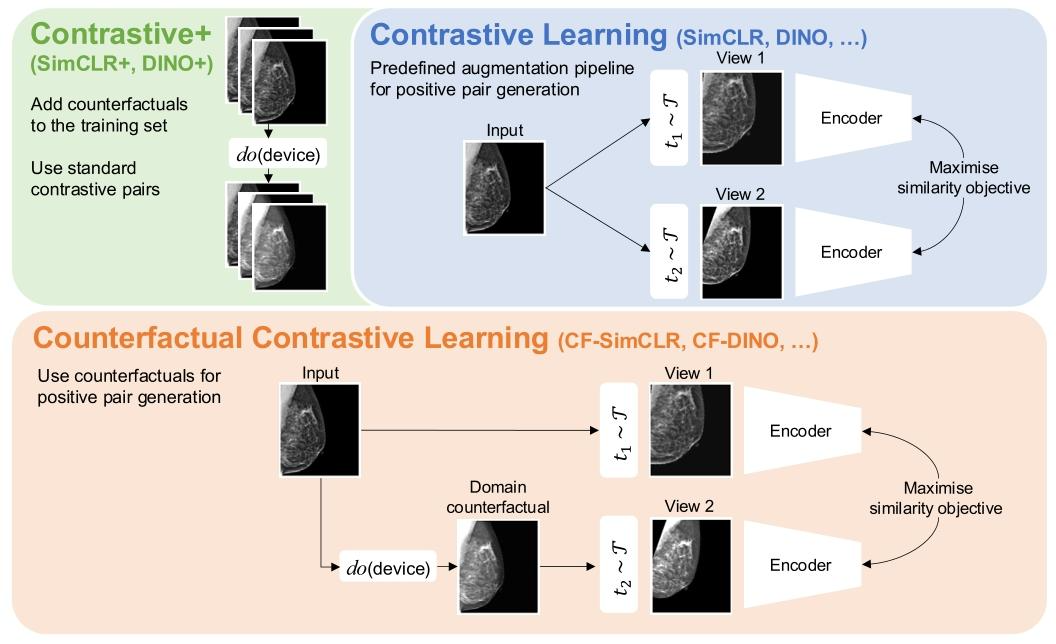
Fig. 1. We propose a novel counterfactual contrastive pair generation framework for improving robustness of contrastively-learned features to distribution shift. As opposed tosolely relying on a pre-defined augmentation generation pipeline (as in standard contrastive learning), we propose to combine real images with their domain counterfactuals tocreate realistic cross-domain positive pairs. Importantly, this proposed approach is independent of the specific contrastive objective employed. The causal image generation modelis represented by the ‘do’ operator. We also compare the proposed method to another approach where we simply extend the training set with the generated counterfactual imageswithout explicit matching with their real counterparts, treating real and counterfactuals as independent training samples.
图1. 我们提出一种新颖的反事实对比样本对生成框架,旨在提高对比学习特征对分布偏移的鲁棒性 与标准对比学习中仅依赖预定义的增强生成流程(\mathcal{T})不同,我们提出将真实图像与其领域反事实图像相结合,以创建真实的跨领域正样本对。重要的是,该方法独立于所采用的特定对比目标。因果图像生成模型由“do”算子表示。我们还将所提方法与另一种方法进行对比:后者仅通过生成的反事实图像扩展训练集,不与真实图像进行显式匹配,而是将真实图像和反事实图像视为独立的训练样本。
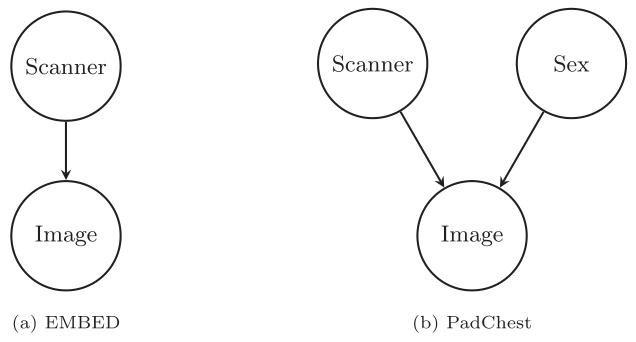
Fig. 2. Causal graphs used to train the counterfactual image generation models usedin this study
图2. 本研究中用于训练反事实图像生成模型的因果图
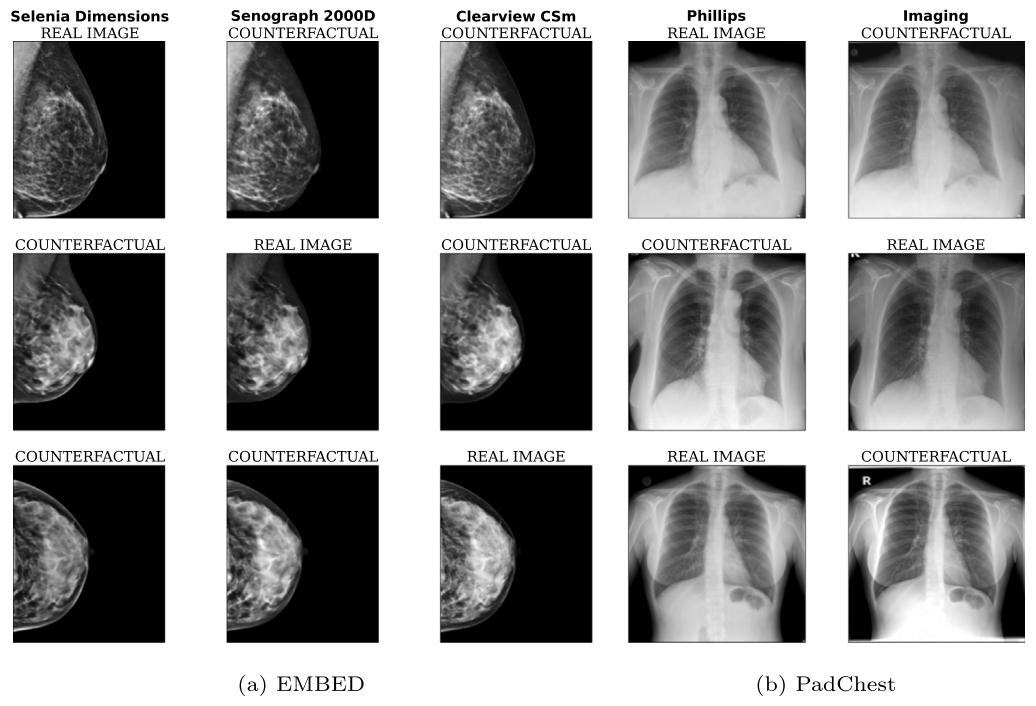
Fig. 3. Examples of counterfactual images generated with our model. Note that on PadChest, text is only imprinted on a subset of Imaging scans (not on Phillips): our modelrespects this by removing text when generating counterfactuals from Imaging to Phillips and vice-versa. Generated images have a resolution of 224 × 224 pixels for PadChest and224 × 192 for EMBED
图3. 利用我们的模型生成的反事实图像示例 注意,在PadChest数据集中,文本仅标记在部分Imaging扫描仪的图像上(未标记在Phillips扫描仪的图像上):我们的模型在从Imaging到Phillips以及从Phillips到Imaging生成反事实图像时,会相应地移除或添加文本,以此遵循这一特性。生成的图像中,PadChest数据集的图像分辨率为224×224像素,EMBED数据集的为224×192像素。
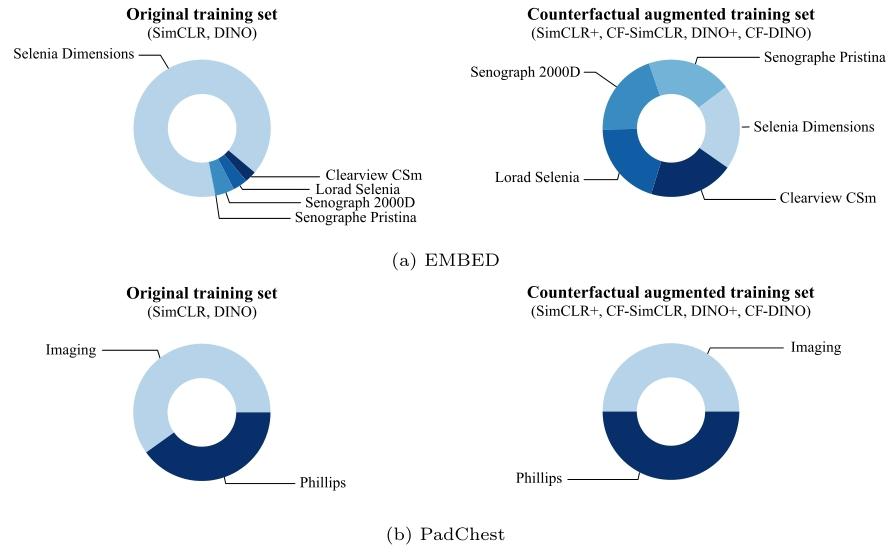
Fig. 4. Distribution of scanners in the original (real-only) training set and the counterfactual-augmented training set for EMBED (top) and PadChest (bottom)
图4. EMBED(上)和PadChest(下)数据集在原始(仅真实图像)训练集与反事实增强训练集中的扫描仪分布情况
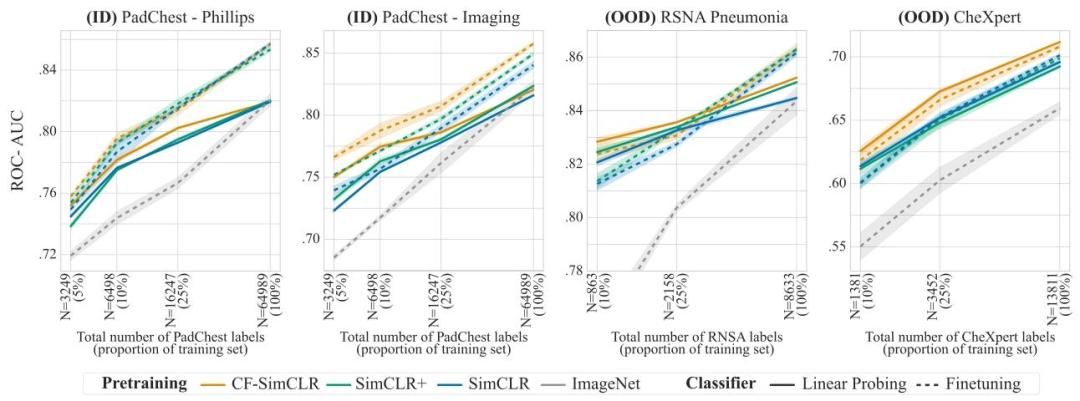
Fig. 5. Pneumonia detection results with linear probing (frozen encoder, solid lines) and finetuning (unfrozen encoder, dashed lines) for models trained with the SimCLR objective.Results are reported as average ROC-AUC over 3 seeds, shaded areas denote ± one standard error. We also compare self-supervised encoders to a supervised baseline initialisedwith ImageNet weights.
图5. 采用SimCLR目标函数训练的模型在肺炎检测任务中的线性探测(编码器冻结,实线)和微调(编码器解冻,虚线)结果 结果以3次随机种子实验的平均ROC-AUC值呈现,阴影区域表示±1标准误差。我们还将自监督编码器与以ImageNet权重初始化的有监督基线模型进行了对比。
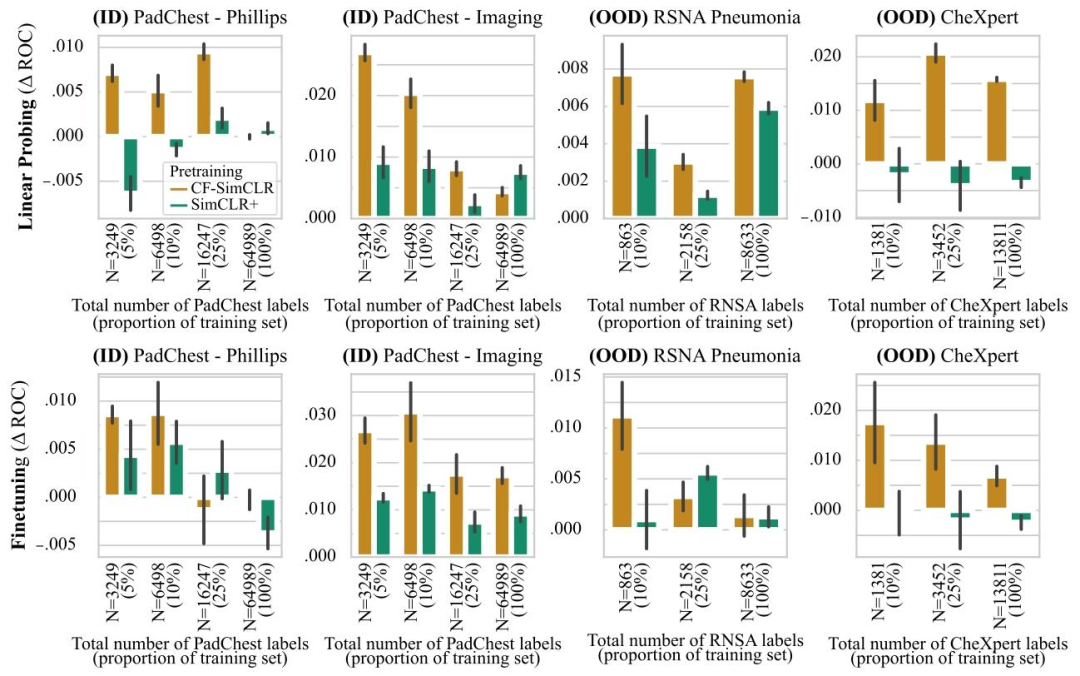
Fig. 6.ROC-AUC difference to SimCLR baseline for CF-SimCLR and SimCLR+ for pneumonia detection. The top row depicts results with linear probing, bottom row shows resultswith model finetuning. Results are reported as average ROC-AUC difference compared to the baseline (SimCLR) over 3 seeds, error bars denote ± one standard error. CF-SimCLRconsistently outperforms encoders trained with standard SimCLR and SimCLR+ (where counterfactuals are added to the training set) for linear probing, and performs best overallfor full model finetuning
图6. CF-SimCLR和SimCLR+相对于SimCLR基线在肺炎检测任务中的ROC-AUC差异** 上排为线性探测结果,下排为模型微调结果。结果以3次随机种子实验中相对于基线(SimCLR)的平均ROC-AUC差异呈现,误差线表示±1标准误差。在 linear probing 中,CF-SimCLR 始终优于采用标准 SimCLR 和 SimCLR+(将反事实图像添加到训练集)训练的编码器;在全模型微调中,CF-SimCLR 总体表现最佳。
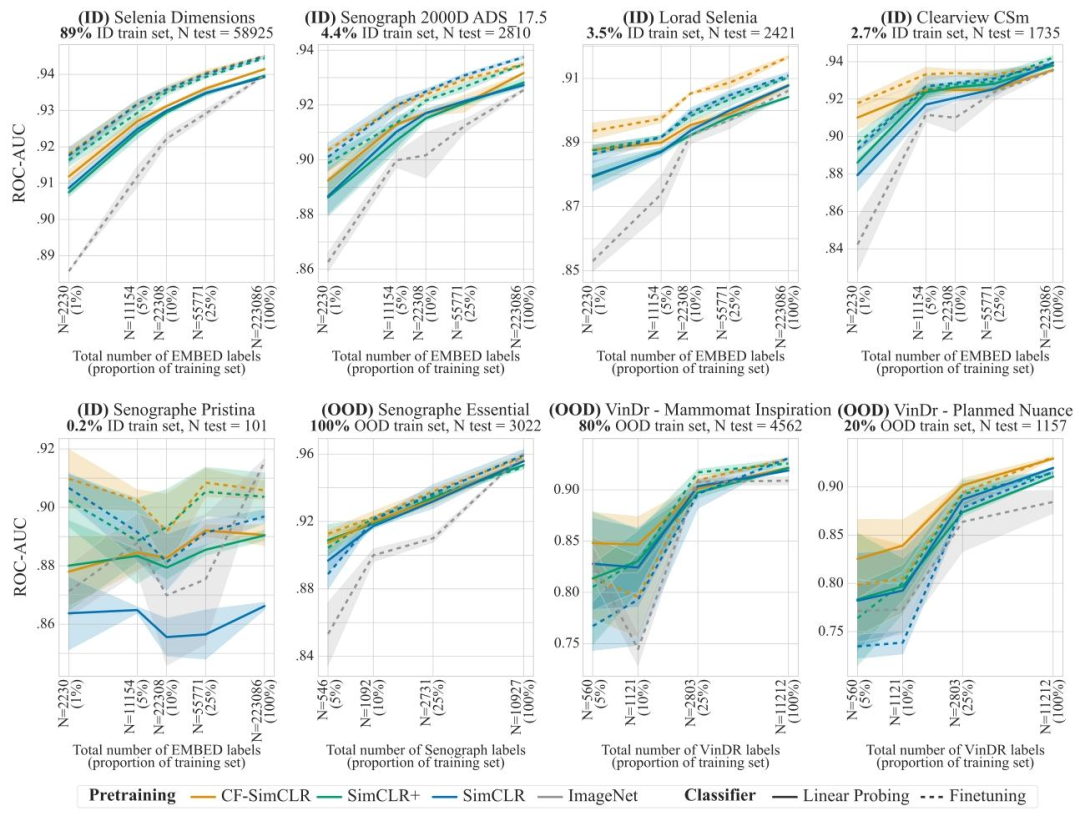
Fig. 7. Breast density results with linear probing (frozen encoder, solid lines) and finetuning (unfrozen encoder, dashed lines) for models trained with SimCLR. Results are reportedas average one-versus-rest macro ROC-AUC over 3 seeds, shaded areas denote ± one standard error. CF-SimCLR performs best overall across ID and OOD data, and improvementsare largest in the low data regime and on under-represented scanners
图7. 采用SimCLR训练的模型在乳腺密度预测任务中的线性探测(编码器冻结,实线)和微调(编码器解冻,虚线)结果 结果以3次随机种子实验的平均“一对多”宏ROC-AUC值呈现,阴影区域表示±1标准误差。CF-SimCLR在分布内(ID)和分布外(OOD)数据上总体表现最佳,且在数据量较少的场景以及针对代表性不足的扫描仪时,改进效果最为显著。
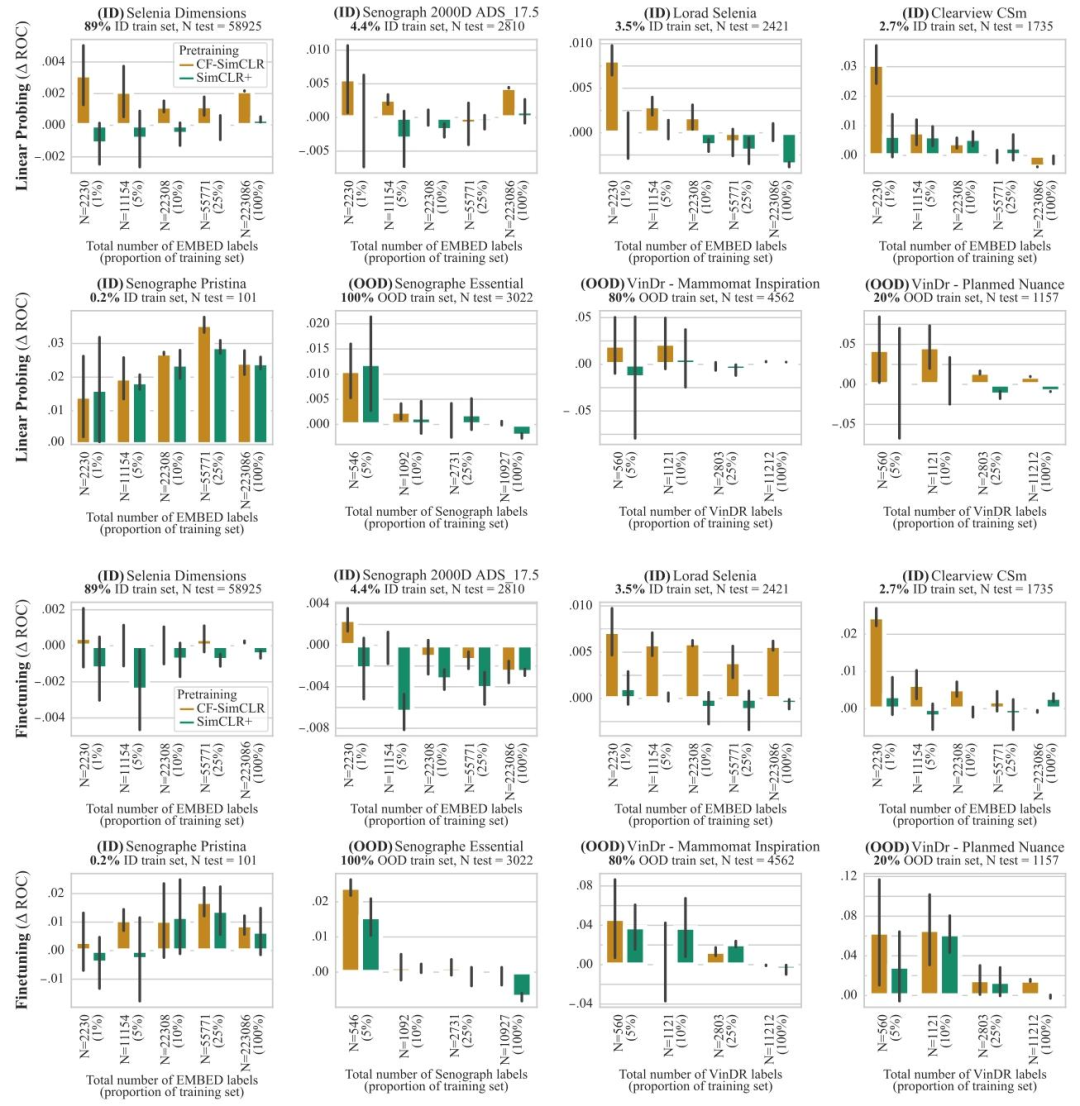
Fig. 8. ROC-AUC difference between SimCLR and CF-SimCLR (resp. SimCLR+) for breast density assessment. The top two rows denote results with linear probing, and the bottomtwo rows show results with model finetuning. Results are reported as average macro ROC-AUC difference compared to the baseline (SimCLR) over 3 seeds, error bars denote ±one standard error. CF-SimCLR overall performs best across ID and OOD data, improvements are largest in the low data regime and on under-represented scanners
图8. SimCLR与CF-SimCLR(分别对应SimCLR+)在乳腺密度评估任务中的ROC-AUC差异 上两行表示线性探测结果,下两行表示模型微调结果。结果以3次随机种子实验中相对于基线(SimCLR)的平均宏ROC-AUC差异呈现,误差线表示±1标准误差。CF-SimCLR在分布内(ID)和分布外(OOD)数据上总体表现最佳,且在数据量较少的场景以及针对代表性不足的扫描仪时,改进效果最为显著。
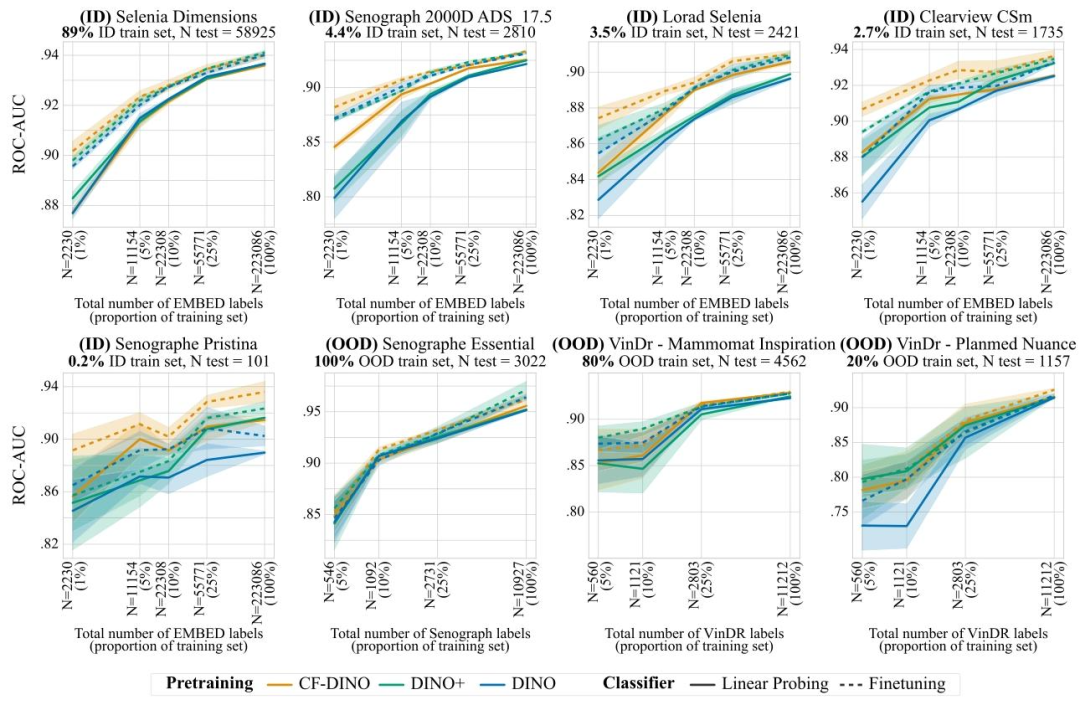
Fig. 9. Breast density classification results for models pretrained with DINO-v2, for both linear probing and finetuning. Results are reported as average one-versus-rest macroROC-AUC over 3 seeds, shaded areas denote ± one standard error. CF-DINO performs best overall, across ID and OOD data, improvements are largest in the low data regime.
图9. 采用DINO-v2预训练的模型在乳腺密度分类任务中的线性探测和微调结果 结果以3次随机种子实验的平均“一对多”宏ROC-AUC值呈现,阴影区域表示±1标准误差。CF-DINO在分布内(ID)和分布外(OOD)数据上总体表现最佳,且在数据量较少的场景中改进效果最为显著。
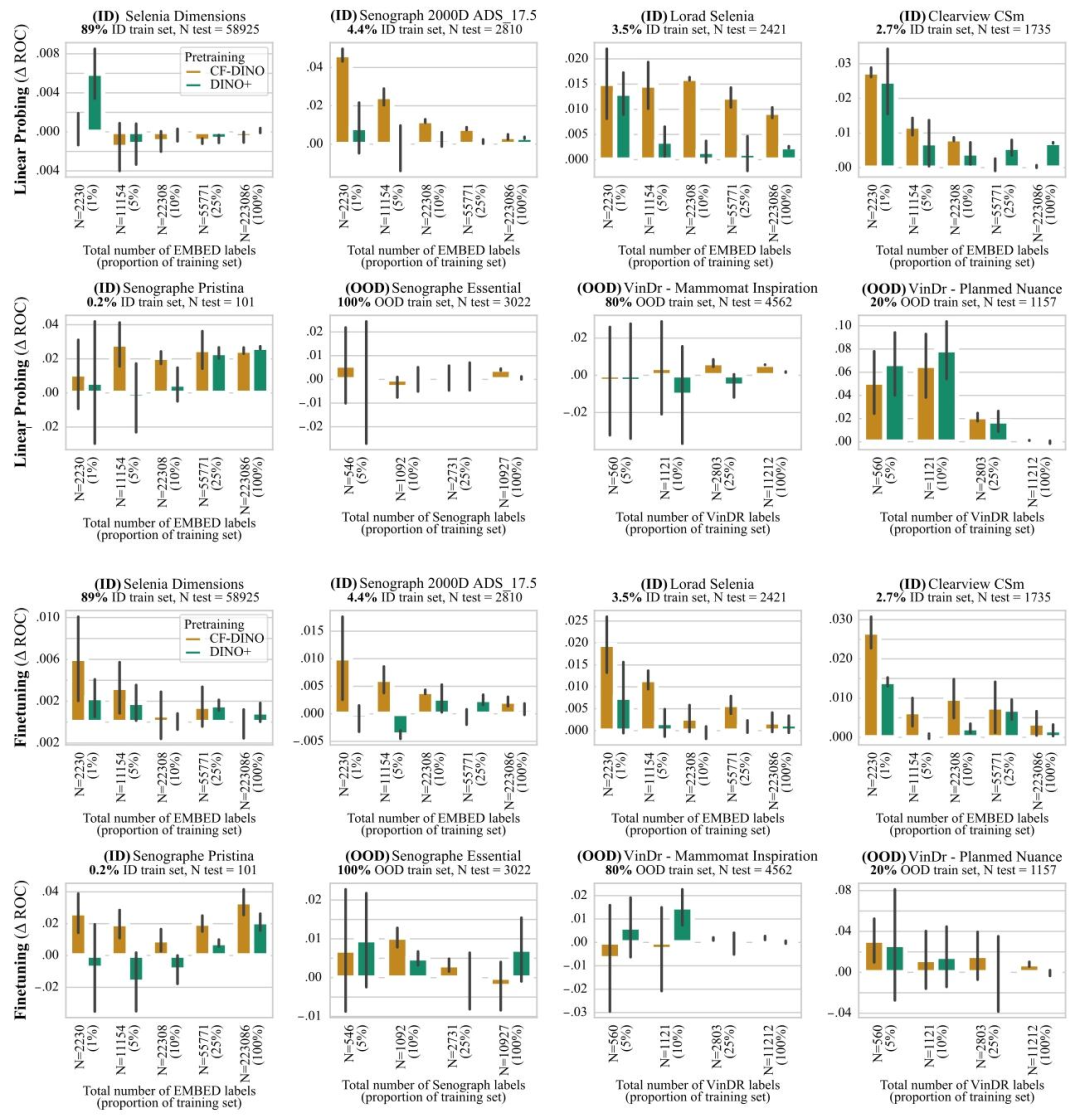
Fig. 10. ROC-AUC difference between DINO and CF-DINO (resp. DINO+). Top two rows denote results with linear probing, bottom two rows results with model finetuning. Resultsare reported as average macro ROC-AUC difference compared to the baseline (DINO) over 3 seeds, error bars denote ± one standard error. CF-DINO overall performs best acrossID and OOD data, improvements are largest in the low data regime and on under-represented scanners.
图10. DINO与CF-DINO(分别对应DINO+)的ROC-AUC差异 上两行表示线性探测结果,下两行表示模型微调结果。结果以3次随机种子实验中相对于基线(DINO)的平均宏ROC-AUC差异呈现,误差线表示±1标准误差。CF-DINO在分布内(ID)和分布外(OOD)数据上总体表现最佳,且在数据量较少的场景以及针对代表性不足的扫描仪时,改进效果最为显著。
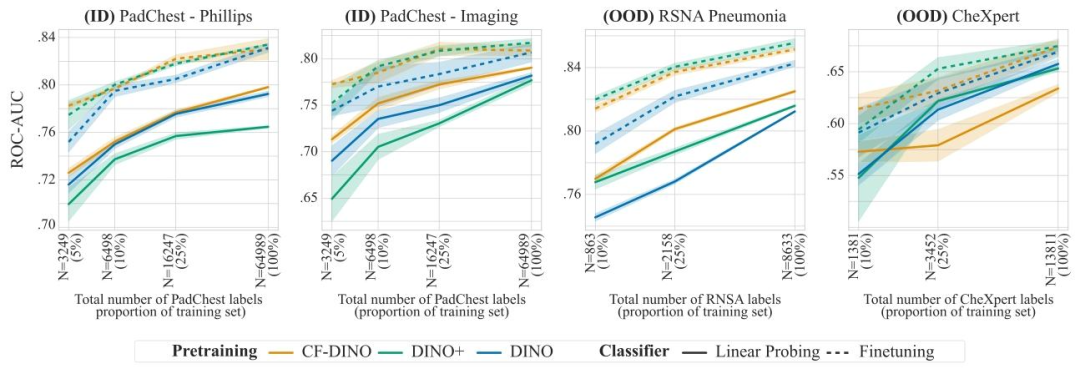
Fig. 11. Pneumonia detection results for models trained with DINO-v2, for both linear probing (frozen encoder) and finetuning. Results are reported as average ROC-AUC over 3seeds, shaded areas denote ± one standard error. CF-DINO consistently outperforms standard DINO.
图11. 采用DINO-v2训练的模型在肺炎检测任务中的线性探测(编码器冻结)和微调结果 结果以3次随机种子实验的平均ROC-AUC值呈现,阴影区域表示±1标准误差。CF-DINO始终优于标准DINO。
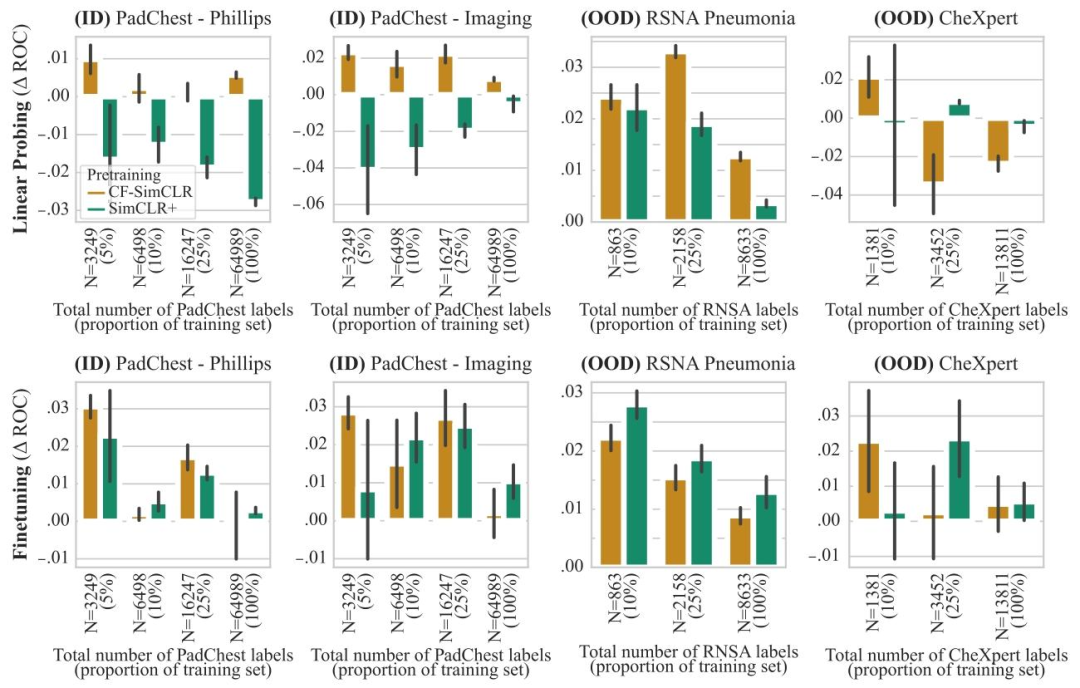
Fig. 12. ROC-AUC difference to DINO baseline for CF-DINO and DINO+ for pneumonia detection. The top row depicts results with linear probing, bottom row show results withmodel finetuning. Results are reported as average ROC-AUC difference compared to the baseline (DINO) over 3 seeds, error bars denote ± one standard error.
图12. CF-DINO和DINO+相对于DINO基线在肺炎检测任务中的ROC-AUC差异 上排为线性探测结果,下排为模型微调结果。结果以3次随机种子实验中相对于基线(DINO)的平均ROC-AUC差异呈现,误差线表示±1标准误差。
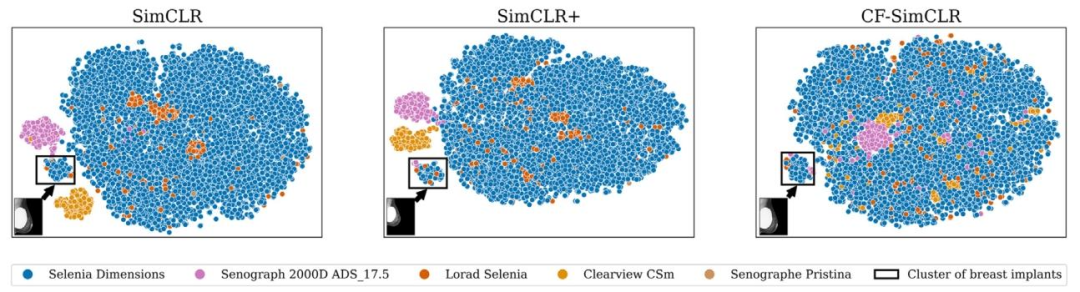
Fig. 13. t-SNE projections of embeddings from 16,000 randomly sampled test images from mammography encoders trained with SimCLR, SimCLR+ and CF-SimCLR. Encoderstrained with SimCLR and SimCLR+ exhibit domain clustering. CF-SimCLR embeddings are substantially less domain-separated and the only disjoint cluster exclusively containsbreasts with implants, semantically different. Thumbnails show a randomly sampled image from each ‘implant’ cluster.
图13. 采用SimCLR、SimCLR+和CF-SimCLR训练的乳腺影像编码器对16,000张随机采样测试图像的嵌入特征的t-SNE投影 经SimCLR和SimCLR+训练的编码器存在领域聚类现象。CF-SimCLR的嵌入特征领域分离度显著降低,且唯一的离散聚类仅包含带有假体的乳腺图像——这在语义上具有明确差异。缩略图展示了每个“假体”聚类中随机采样的图像。
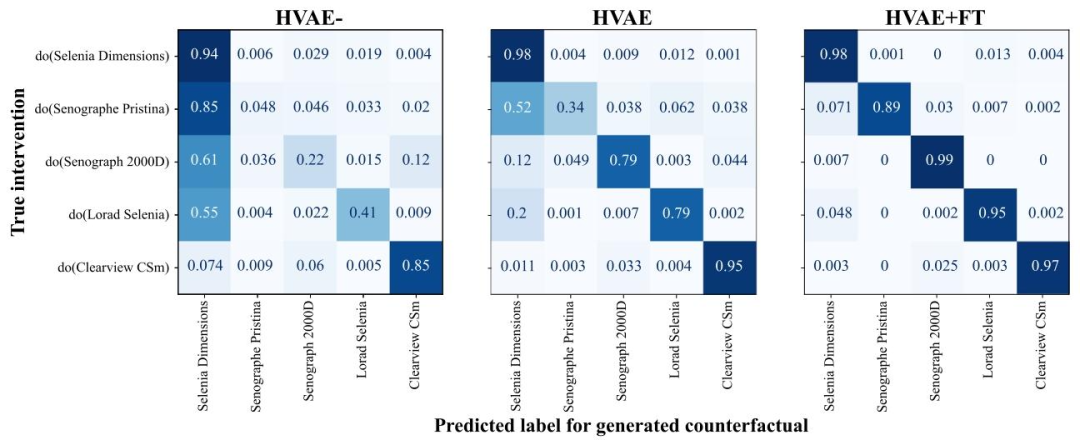
Fig. 14. Effectiveness comparison for the three counterfactual models considered in this ablation study, by intervention. Computed on 8304 validation set samples.
图 14. 本消融研究中所考虑的三种反事实模型在不同干预条件下的有效性对比基于 8304 个验证集样本计算得出。
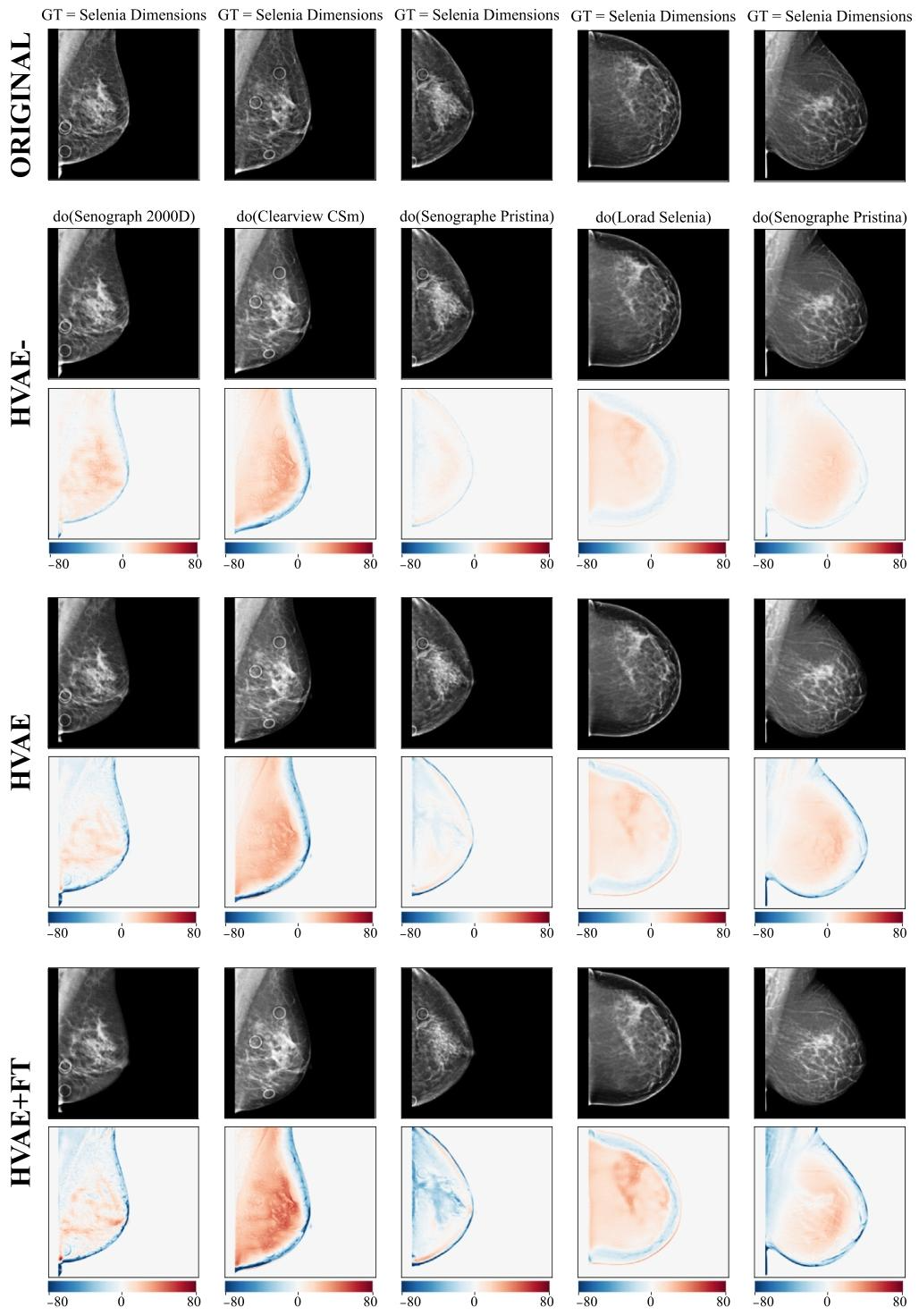
Fig. 15. Qualitative of comparison of the three counterfactual generation models, HVAE-, HVAE and HVAE+FT compared in the ablation study. For each model we show generatedcounterfactuals as well as direct effect maps. Direct effects give a visual depiction of the increase in effectiveness across the three models from top to bottom. We also observethat all models preserve semantic identity very well, a key aspect in positive pair creation contrastive learning
图15. 消融研究中三种反事实生成模型(HVAE⁻、HVAE和HVAE+FT)的定性对比 对于每个模型,我们展示了生成的反事实图像以及直接效应图。直接效应图直观呈现了从顶部到底部三个模型有效性的提升。我们还观察到,所有模型都能很好地保留语义一致性——这是对比学习中构建正样本对的关键要素。
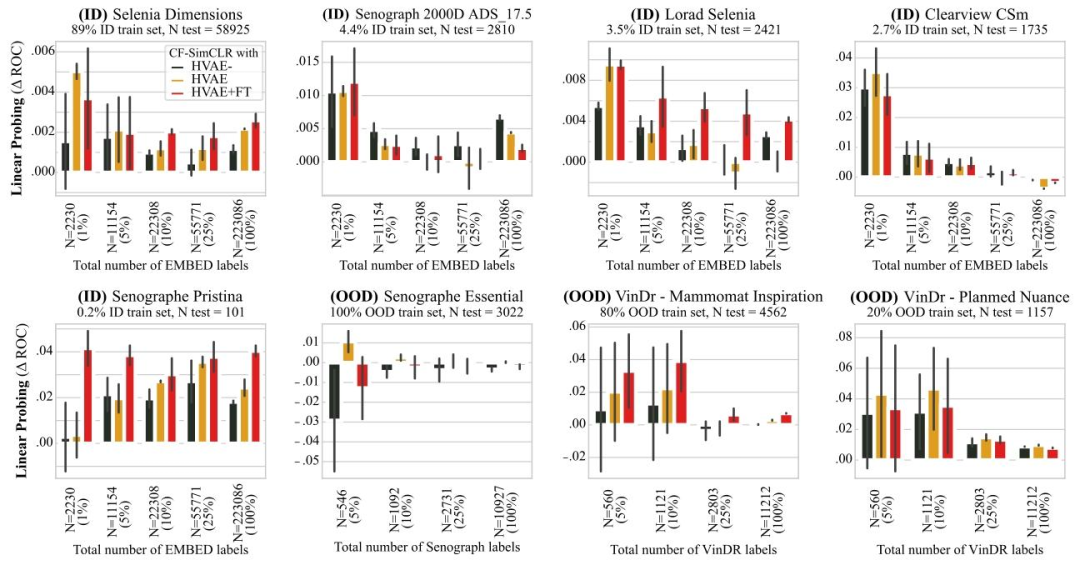
Fig. 16. Effect of counterfactual quality on downstream performance. Results are reported as average macro ROC-AUC difference compared to the baseline (SimCLR) over 3 seedsfor linear probing, error bars denote ± one standard error. We compare running CF-SimCLR with (i) HVAE- a counterfactual generation model of lesser effectiveness, (ii) HVAEthe generation model used in the rest of this study, (iii) HVAE+FT a counterfactual generation model with higher effectivenes
图16. 反事实质量对下游任务性能的影响 结果以3次随机种子实验中线性探测相对于基线(SimCLR)的平均宏ROC-AUC差异呈现,误差线表示±1标准误差。我们对比了使用以下三种反事实生成模型的CF-SimCLR性能:(i)HVAE⁻——一种有效性较低的反事实生成模型;(ii)HVAE——本研究其余部分所使用的生成模型;(iii)HVAE+FT——一种有效性更高的反事实生成模型。
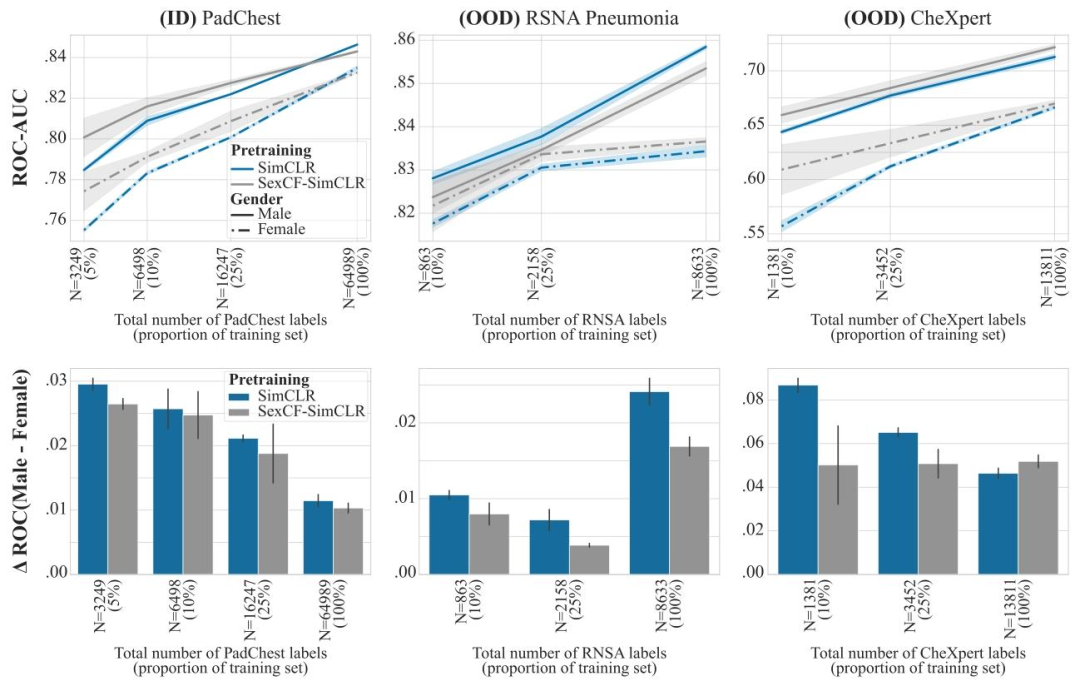
Fig. 17. Improving sub-group performance with counterfactual contrastive learning. Pneumonia detection results with linear probing for encoders trained with SimCLR andSexCF-SimCLR. In SexCF-SimCLR, we generate sex counterfactuals instead of domain counterfactuals for positive pair generation to improve robustness to subgroup shift and,ultimately, performance on under-represented subgroups. Top row: performance for the male (solid line) and female (dashed line) subgroups, reported as average ROC-AUC over3 seeds, shaded areas denote ± standard error. Bottom row: performance disparities across the two subgroups, reported as average ROC-AUC difference between the male andfemale subgroups, over 3 seeds. Sex CF-SimCLR reduces sub-group disparities for all datasets, substantially increasing performance on the female sub-group when limited amountsof labels are available, both on ID and OOD datasets
图17. 利用反事实对比学习提升亚组性能 采用SimCLR和SexCF-SimCLR训练的编码器在肺炎检测任务中的线性探测结果。在SexCF-SimCLR中,我们生成性别反事实而非领域反事实用于正样本对构建,以增强对亚组偏移的鲁棒性,并最终提升代表性不足亚组的性能。 上排:男性(实线)和女性(虚线)亚组的性能,以3次随机种子实验的平均ROC-AUC值呈现,阴影区域表示±标准误差。 下排:两个亚组间的性能差异,以3次随机种子实验中男性与女性亚组的平均ROC-AUC差值呈现。 SexCF-SimCLR降低了所有数据集的亚组性能差异,在标注数据有限的情况下,显著提升了女性亚组在分布内(ID)和分布外(OOD)数据集上的性能。
Table
表
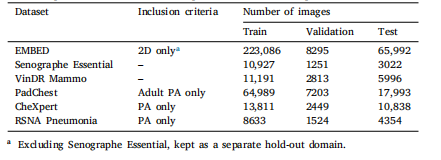
Table 1Datasets splits and inclusion criteria. Splits are created at the patient level
表1 数据集划分及纳入标准 数据集划分以患者为单位进行。
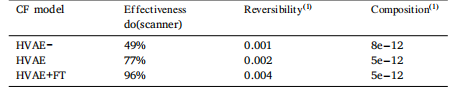
Table 2Axiomatic soundness metrics (Monteiro et al., 2022) for the three models consideredin this ablation study. Computed on the 8304 validation set samples
表2 本消融研究中所考虑的三种模型的公理合理性指标(Monteiro等人,2022) 基于8304个验证集样本计算得出。
:朴素贝叶斯分类算法)



、主机虚拟机(Windows、Ubuntu)、手机(笔记本热点),三者进行相互ping通)














