华大时空组学空转图像处理
library(png)
library(tiff)
st <- readRDS('01.Stereo-seq/output_all/Demo_Mouse_Kidney/outs/feature_expression/seurat_out.rds')
dim(st@assays$Spatial@counts)
st@assays$Spatial@counts[1:4,1:4]
coord.df <- data.frame(imagerow = st$x, imagecol=st$y,nCount_Spatial = st$nCount_Spatial,row.names = st$X_index)
min(coord.df$imagerow);max(coord.df$imagerow);min(coord.df$imagecol);max(coord.df$imagecol)
image <- readTIFF("/biodata2/08.user/zhengyzh/01.Stereo-seq/output_all/Demo_Mouse_Kidney/outs/image/bin1_img_tissue_cut.tif")
用于seurat
st@images$slice1 = new(Class = 'SlideSeq',assay = "Spatial",key = "image_",coordinates = coord.df
)
图像分bin转化
bin_matrix_fast <- function(mat, bin_size=200) {n <- nrow(mat)m <- ncol(mat)trimmed_n <- floor(n / bin_size) * bin_sizetrimmed_m <- floor(m / bin_size) * bin_sizemat <- mat[1:trimmed_n, 1:trimmed_m]dim(mat) <- c(bin_size, trimmed_n/bin_size, bin_size, trimmed_m/bin_size)result <- apply(mat, c(2, 4), mean, na.rm = TRUE)return(result)
}img <- bin_matrix_fast(image, 50)
img <- img[dim(img)[1]:1,]
ggplot2手搓实现切片图像背景图效果
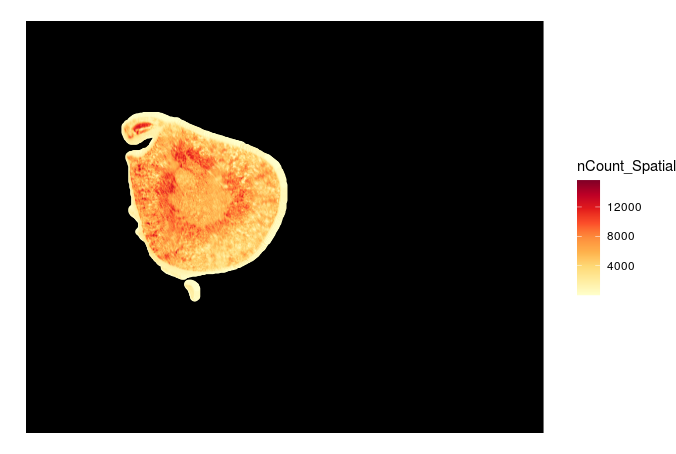
ggplot() +annotation_raster(img, 0, dim(img)[1], 0, dim(img)[2]) + geom_point(data = coord.df, aes(x=imagerow/50,y=imagecol/50,color=nCount_Spatial)) + scale_color_gradientn(colours = RColorBrewer::brewer.pal(9,'YlOrRd')) +xlim(0,dim(img)[1]) + ylim(0,dim(img)[2]) +theme_void()
gray_img <- 0.299 * img[,,1] + 0.587 * img[,,2] + 0.114 * img[,,3]
grid.raster(gray_img, interpola = TRUE)
gray_array <- array(gray_img, dim = c(dim(img)[1], dim(img)[2], 1))






![【题解 | 两种做法】洛谷 P4208 [JSOI2008] 最小生成树计数 [矩阵树/枚举]](http://pic.xiahunao.cn/【题解 | 两种做法】洛谷 P4208 [JSOI2008] 最小生成树计数 [矩阵树/枚举])









+ AI:智能工业的未来趋势全解析)



